Question from a user:
I´d like to extract a list of all marine fish species (e.g., within the Subclass Actinopterygii) with occurrences inside a number of polygons (marine ecoregions). Is there an easy way of doing this?
GBIF has a geometry parameter in their search API, that accepts well known text polygon. If you need to search many polygons, do one search for each.
library("rgbif")
poly1 <- "POLYGON((-77.42 34.19,-74.43 36.35,-72.32 39.81,-67.93 42.19,-66.87 40.34,-73.90 33.46,-77.42 34.19))"
key <- name_backbone(name = "Actinopterygii", kingdom = "Animals")$usageKey
(res <- occ_search(taxonKey = key, geometry = poly1, hasCoordinate = TRUE))
Records found [100247]
Records returned [500]
No. unique hierarchies [273]
No. media records [10]
Args [taxonKey=204, hasCoordinate=TRUE, geometry=POLYGON((-77.42 34.19,-74.43
36.35,-72.32 39.81,-67.93 42.19,-66.87 40.34,-73.90 33.46,-77.42 34.19)),
limit=500, offset=0, fields=all]
First 10 rows of data
name key decimalLatitude decimalLongitude
1 Prionotus carolinus 1098922649 34.18340 -76.60000
2 Sphyraena barracuda 1098922679 34.26690 -76.63350
3 Centropyge argi 1098922662 34.18340 -76.60000
4 Balistes capriscus 1098922668 34.18340 -76.60000
5 Paralichthys lethostigma 1098922650 34.18340 -76.60000
6 Lipophrys pholis 1098922671 34.18340 -76.60000
7 Pomacanthus arcuatus 1098922691 34.18340 -76.60000
8 Seriola dumerili 1098922666 34.18340 -76.60000
9 Rachycentron canadum 1098920153 35.09541 -75.71921
10 Selar crumenophthalmus 1123777329 34.27800 -76.64500
.. ... ... ... ...
Variables not shown: issues (chr), datasetKey (chr), publishingOrgKey (chr),
publishingCountry (chr), protocol (chr), lastCrawled (chr), lastParsed (chr),
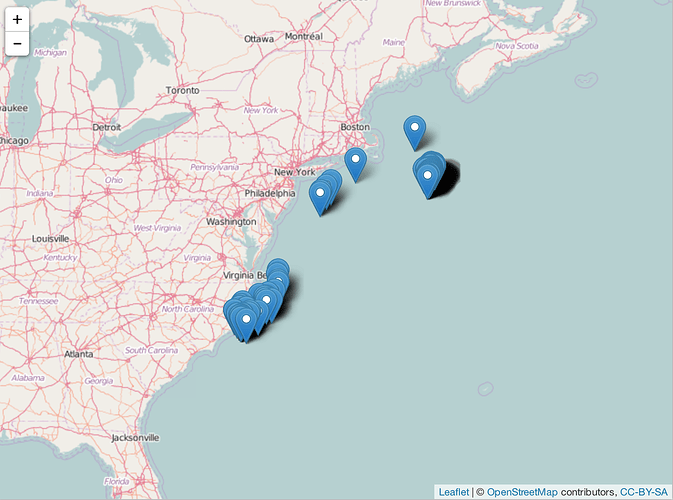
Plot to make sure
Polygon searched
library("geojsonio")
library("lawn")
library("dplyr")
res$data %>%
select(name, decimalLatitude, decimalLongitude) %>%
rename(latitude = decimalLatitude, longitude = decimalLongitude) %>%
geojsonio::geojson_json() %>%
lawn::view()
Last, if you need a lot of data, e.g., more than 200,000 records, use the GBIF download API. If you need that, I can show some examples for that.
Get a species list
Depends on what exactly you want, but the simplest form is to just get a unique
list of species (using the data above)
splist <- unique(res$data$name)
splist[1:5]
[1] "Prionotus carolinus" "Sphyraena barracuda" "Centropyge argi"
[4] "Balistes capriscus" "Paralichthys lethostigma"
If there’s enough interest we could maybe add a helper function to rgbif to extract species lists, but it’s super simple to do on your own, and there’s a variety of columns you could pull out for the names.
Pass in a shapefile?
Not in rgbif, but in spocc. And not a shapefile directly, but convert to a spatial class first (one of SpatialPolygons or SpatialPolygonsDataFrame), then pass into the search function in spocc, which is occ(), similar to occ_search() in rgbif. An example:
library("spocc")
library("sp")
library("maptools")
Single polygon in SpatialPolygons class
one <- Polygon(cbind(c(91,90,90,91), c(30,30,32,30)))
spone = Polygons(list(one), "s1")
sppoly = SpatialPolygons(list(spone), as.integer(1))
out <- occ(geometry = sppoly, from = "gbif", limit=5)
out$gbif
Geometry [<geo1> (5)]
name longitude latitude prov issues key
1 Falco cherrug 90.6781 30.2668 gbif gass84 959430655
2 Ptyonoprogne rupestris 90.6781 30.2668 gbif gass84 959430642
3 Phoenicurus fuliginosus 90.6781 30.2668 gbif gass84 959431391
4 Montifringilla ruficollis 90.6781 30.2668 gbif gass84 959430887
5 Phoenicurus ochruros 90.6781 30.2668 gbif gass84 959430681
Variables not shown: datasetKey (chr), publishingOrgKey (chr), publishingCountry
(chr), protocol (chr), lastCrawled (chr), lastParsed (chr), extensions (chr),
...
From a shapefile
xx <- readShapeSpatial(system.file("shapes/sids.shp", package="maptools")[1],
IDvar="FIPSNO", proj4string=CRS("+proj=longlat +ellps=clrk66"))
poly <- SpatialPolygons(list(xx@polygons[[1]]), 1L) # just get one of the polygons for brevity
out <- occ(geometry = poly, from = "gbif", limit=5)
out$gbif
Geometry [<geo1> (5)]
name longitude latitude prov issues key
1 Daucus carota -79.44724 36.14592 gbif cdround,cudc,gass84 1098912986
2 Taraxacum croceum -79.46419 36.01529 gbif cdround,cudc,gass84 1211970689
3 Trifolium pratense -79.43243 36.07145 gbif cdround,cudc,gass84 1098914121
4 Photinus pyralis -79.40369 36.04958 gbif cdround,cudc,gass84 1143519750
5 Phytolacca americana -79.49715 36.17032 gbif cdround,cudc,gass84 1132405750
Variables not shown: datasetKey (chr), publishingOrgKey (chr), publishingCountry
(chr), protocol (chr), lastCrawled (chr), lastParsed (chr), extensions (chr),
...